
qScript cDNA SuperMix
- Stabilized 5X concentrated master mix minimizes pipetting to significantly improve assay accuracy
- Sensitive first-strand cDNA synthesis of RNA sequences ≤1kb for quantitative and conventional two-step RT-PCR
- Broad linear dynamic detection range with limiting (10pg) or plentiful (1µg) samples of total RNA
- Pre-blended with ribonuclease inhibitor protein (RIP) to prevent RNA template degradation during incubation and a proprietary blend of random and oligo(dT) primers, optimized to deliver unbiased representation of 5’ and 3’ sequences
qScript cDNA SuperMix is intended for molecular biology applications. This product is not intended for the diagnosis, prevention or treatment of a disease.
qScript cDNA SuperMix
Description
qScript cDNA SuperMix is a sensitive and easy-to-use 1-tube reagent for first-strand cDNA synthesis that combines a highly-modified RNAse H+ mutant of M-MLV together with ribonuclease inhibitor protein (RIP) in a rigorously optimized formulation for real-time qPCR applications. The stabilized SuperMix formulation has been rigorously optimized to deliver sensitive, linear assay performance across a spectrum of relative abundance and input RNA (10pg - 1ug). qScript cDNA SuperMix reagent performance is unaffected by repetitive freeze/thaw cycling (>20X), conferring greater ease-of-use and consistent assay performance. Oligo (dT) and random primers are pre-blended in a precise ratio to provide equal representation of 5' and 3'-sequences for accurate gene expression quantification. For gene-specific priming (GSP) or two-step RT-PCR of RNA exceeding 1kb total length, see our qScript Flex cDNA Kit.
qScript cDNA SuperMix is intended for molecular biology applications. This product is not intended for the diagnosis, prevention or treatment of a disease.
Details
Single-tube, 5X concentrated reagent containing:
- Reaction buffer with optimized concentrations of molecular-grade MgCl2, dATP, dCTP, dGTP, dTTP
- Recombinant ribonuclease inhibitor protein (RIP)
- qScript reverse transcriptase
- Titrated concentrations of random hexamer and oligo(dT) primer
- Proprietary enzyme stabilizers and performance-enhancing additives
Performance Data
Protocol Comparison
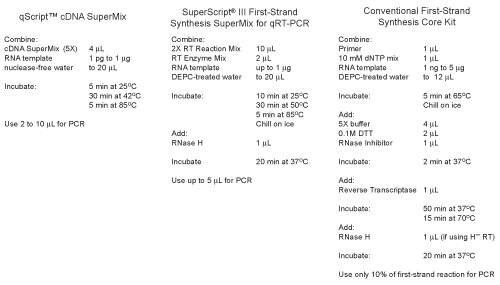
Easy-to-use cDNA SuperMix format
Higher Yield (more accurate representation) of low abundance gene with qScript - PP2A
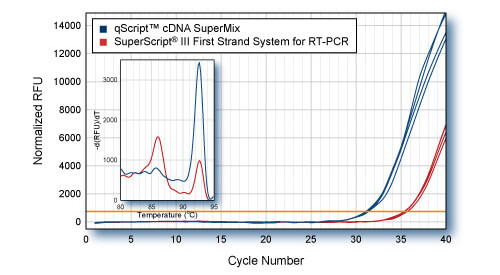
First-strand synthesis was carried out using 1 μg HeLa cell total RNA with either qScript cDNA SuperMix or SuperScript® III First-Strand Synthesis System for RT-PCR. 5 ng total RNA equivalent (1/200th of each cDNA reaction) was used for SYBR Green qPCR of PP2A gene with PerfeCTa SYBR Green SuperMix. Inset: Dissociation (melt curve) analysis of SYBR Green qRT-PCR products shows amplification of correct amplicon in qScript reactions. Lower cDNA yield from SuperScript III reactions results in amplification of non-specific product.
Sensitivity - TRRAP comparison to SuperScript III SuperMix First Strand SuperMix for qPCR & QuantiTect Reverse Transcription Kit
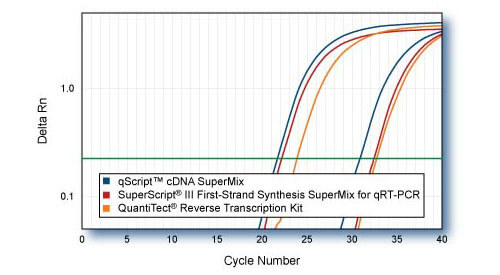
First-strand cDNAs were synthesized following each manufacturers protocol with 1 μ or 1 ng of HeLa cell total RNA as template. 1/10th of first-strand reactions were used for qRT-PCR of 5′-end region of TRRAP gene with PerfeCTa SYBR Green SuperMix. Averaged plots from 4 replicate qPCR reactions are shown.
Reproducibility - CDKN1B
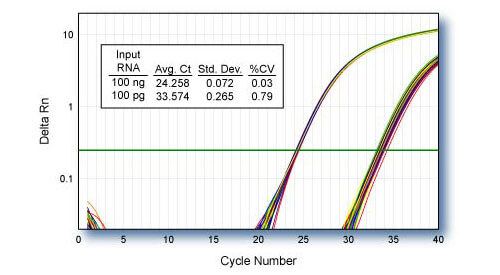
qScript cDNA SuperMix was used for 48 independent first-strand reactions containing 100 ng or 100 pg of HeLa cell total RNA template. 1/10th of each first-strand reaction was used for TaqMan® qPCR of CDKN1B using PerfeCTa qPCR SuperMix.
Linearity - ACTB
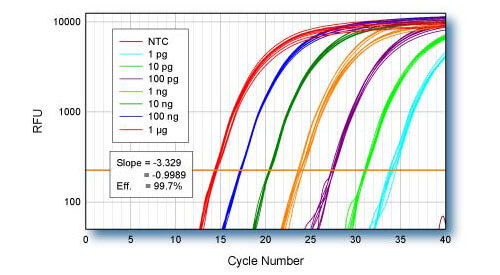
qScript cDNA Supermix was used for first-strand cDNA synthesis using log-fold serial dilutions of HeLa cell total RNA from 1 μg to 1 pg. Eight replicate cDNA reactions were performed for each input quantity of RNA. 1/10th of each first-strand reaction was used for qPCR of the ACTB gene using PerfeCTa SYBR Green SuperMix.
Resources
Flyers
Product Manuals
Application Notes
CofA (PSFs)
SDSs
Publications
Customer Product Reviews
| 5 star | 100% | |
| 4 star | 0% | |
| 3 star | 0% | |
| 2 star | 0% | |
| 1 star | 0% |
 qScript cDNA SuperMix
qScript cDNA SuperMix

Performed comparably with a quicker runtime and a lower price point compared to our previous cDNA synthesis kit.
It was good but not outperforming
Product works well and is up to our standards.