AccuStart II PCR ToughMix
- Stabilized 2X PCR SuperMix enables convenient room-temperature setup and is unaffected by repetitive freeze-thaw
- High-yielding, ultrapure modified Taq DNA polymerase delivers robust, reliable duplex assay performance
- Stringent, ultrapure antibody hotstart ensures sensitive and specific target amplification
- Separate electrophoretic mobility dye reduces risk of post-PCR cross contamination with gel electrophoresis
AccuStart II PCR ToughMix is intended for molecular biology applications. This product is not intended for the diagnosis, prevention or treatment of a disease.
AccuStart II PCR ToughMix
Description
AccuStart II PCR ToughMix is a 2X concentrated ready-to-use reaction cocktail for PCR amplification of DNA templates that overcomes many known inhibitors of PCR often present in crude samples extracted from environmental specimens, plant tissues, or animal tissues. The only user supplied components are DNA template and molecular grade water.
A key component to AccuStart PCR ToughMix is an ultra pure, highly processive thermostable DNA polymerase that is combined with high avidity monoclonal antibodies. These antibodies bind the polymerase and keep it inactive prior to the initial PCR denaturation step. This enables specific and efficient primer extension with the convenience of room temperature reaction assembly. Similar to Taq DNA polymerase, the activated polymerase in AccuStart II PCR ToughMix possesses 5'>3' DNA polymerase activity and a double-strand specific 5'>3' exonuclease. The polymerase does not have 3'-exonuclease activity and is free of any contaminating endo or exonuclease activities. PCR products generally contain non-templated dA additions and can be cloned using vectors that have a single 3'-overhanging thymine residue on each end.
GelTrack Loading Dye is a mixture of blue and yellow electrophoresis-tracking dyes that migrate at approximately 4kb and 50 bp. This optional component simplifies post-PCR analysis with gel electrophoresis and eliminates potential for cross contamination by enabling direct transfer of PCR products to the gel sample wells.
The GelTrack Loading Dye solution is not included with the sample size 95142-100.
Details
- 2X concentrated PCR SuperMix containing reaction buffer with optimized concentrations of molecular-grade MgCl2, dNTPs, AccuStart II hot start Taq DNA polymerase, ToughMix additives and stabilizers.
- GelTrack Loading Dye: 50X concentrated mixture blue and yellow electrophoresis-tracking dyes. (Does not contain a density reagent.)
The GelTrack Loading Dye solution is not included with the sample size 95142-100.
Performance Data
Inhibitor Resistance of AccuStart II PCR ToughMix: PCR in the presence of polyphenol spike
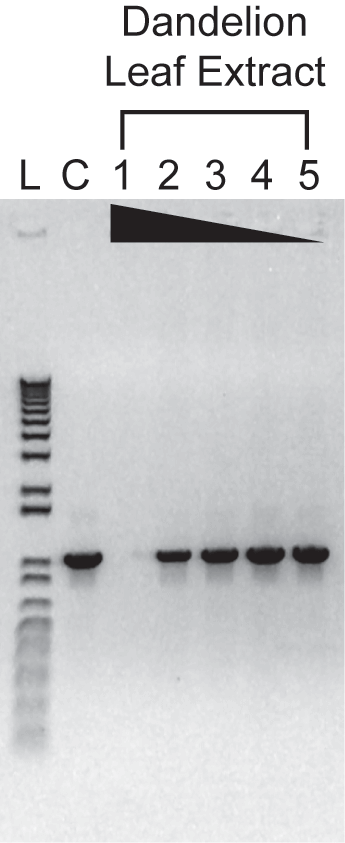
Varying amounts of a polyphenol-rich plant extract (0.2, 0.06, 0.02, 0.006, or 0.002 uL) were added to 25-uL PCRs containing 10,000 copies of a control template. Amplification was carried out for30 cycles of: 94°C, 15s; 60°C, 20s; 72°C, 1 min. 1/5th of each reaction was analyzed on a 01%agarose, 0.5X TBE gel containing 0.25 mg/mL ethidium bromide. As little as 0.002 uL of the crude plant lysate inhibited control reactions with a conventional PCR master mix (data not shown).
Inhibitor Resistance of AccuStart II PCR ToughMix
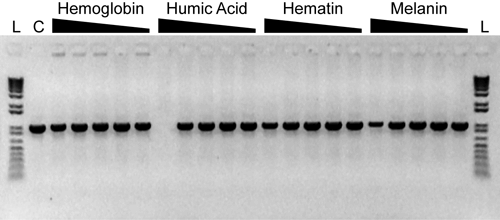
A 1-kb fragment from 1e4 copies of the Tetracyclin resistance gene was amplified in 20-µL reaction volumes according to the recommended protocol. Reactions were challenged with varying concentrations of different PCR inibitors as summarized below. Following a 3 min activation at 94°C; PCR was for 30 cycles of: 94°C, 15s; 60°C, 20s; 72°C, 1 min. 1/5th of each reaction was analyzed on a 01% agarose, 0.5X TBE gel conatining 0.25 mg/mL ethidium bromide.
Hemoglobin: 316 ng/µL, 100 ng/µL, 31.6 ng/µL, 10 ng/µL, 3.16 ng/µL
Humic Acid: 31.6 ng/µL, 10 ng/µL, 3.16 ng/µL, 1 ng/µL, 0.316 ng/µL
Hematin: 100 µM, 31.6 µM, 10 µM, 3.16 µM, 1 µM
Melanin: 10 ng/µL, 3.16 ng/µL, 1 ng/µL, 0.316 ng/µL, 0.1 ng/µL
C: control reactions without inhibitor
L: 1 Kb Plus DNA Ladder (Invitrogen)
Resources
Flyers
Product Manuals
Application Notes
CofA (PSFs)
SDSs
Publications
Customer Product Reviews
| 5 star | 60% | |
| 4 star | 20% | |
| 3 star | 0% | |
| 2 star | 20% | |
| 1 star | 0% |
 AccuStart II PCR ToughMix
AccuStart II PCR ToughMix

This works well for SSR multiplexing! An easy 1-tube solution to PCRing.
Very Good for colony PCR !
I was really hoping this mastermix would work for my pcr. The hot start specification would have allowed me to work on the benchtop. Even though your product could endure multiple freeze-thaw cycles, I aliquoted working vials to avoid freeze-thaw just in case. I followed your product recommendation for my first few tests of this mastermix and the Extracta DBS. I got very weak target amplification for 2 out 6 samples (samples were run before and results are known) and very dark smearing. Upon further reading of the FTA cards I am working with (FTA Classic) they recommend not to elude DNA from the cards and these cards are different than Gutherie cards as they have proprietary lysing agents and preservation chemicals stabilize DNA and recommend washing with their proprietary rinse buffer. I switched to washing with their rinse buffer and direct pcr amplification using your ToughMix mastermix with moderate success. I was able to get nice target amplification on 1 round of tests on the same 6 samples but have not been able to replicate those results. Inconsistent results with either no target bands at all or very faint band for 1 or 2 samples. I then tried washing with the FTA rinse buffer and extracting DNA with your Extracta DBS with no target amplification. I exhausted the sample of mastermix you provided. I think if I had more time and more mastermix I would have contacted you for information on the MgCl2 concentration as I believe I may have been able to get direct amplification of my target amplicon by adjusting the [MgCl2]. In the end I used a colleagues mastermix to test and was able to get reproducible direct amplification of my target.
Sorry to hear this didn’t perform as you had hoped for these applications. A member from the applications team has reached out to provide additional support for you and your team.