PerfeCTa qPCR ToughMix
- ToughMix additives neutralize PCR inhibitors to ensure reliable assay performance with a spectrum of starting materials including: clinical specimens, plants, soil, environmental or complex food matrices
- Superior assay sensitivity and target precision with AccuStart™ II enzyme technology – maximum-yielding, engineered Taq DNA polymerase with stringent, ultrapure antibody hotstart
- Easy-to-use 2X concentrated master mixes with AccuVue™ plate loading dye and optimized passive reference dye for simplified reaction setup
- Supports efficient vortex mixing with proprietary anti-foaming technology
- Consistent and reliable performance with exceptional reagent stability (uninhibited after 20X freeze-thaw or 30 days at 22°C)
PerfeCTa qPCR ToughMix is intended for molecular biology applications. This product is not intended for the diagnosis, prevention or treatment of a disease.
PerfeCTa qPCR ToughMix
PerfeCTa qPCR ToughMix ROX
PerfeCTa qPCR ToughMix Low ROX
PerfeCTa qPCR ToughMix UNG
PerfeCTa qPCR ToughMix UNG ROX
PerfeCTa qPCR ToughMix UNG Low ROX
Description
PerfeCTa qPCR ToughMix is a 2X concentrated ready-to-use reaction cocktail for PCR amplification of DNA templates that relieves several types of PCR inhibition commonly encountered with crude extracts, environmental specimens, plant tissues, animal tissues, and complex food matrices. This robust real-time qPCR reagent provides maximum sensitivity and PCR efficiency with a variety of fluorogenic probe chemistries, including TaqMan® hydrolysis probes. The only user-supplied components are primers, probe(s), and DNA template. Pre-blended with inert AccuVue plate loading dye to help minimize pipette errors during setup and provides visual confirmation of thorough mixing.
A key component of PerfeCTa qPCR ToughMix is an ultra pure, highly processive thermostable DNA polymerase that is combined with high avidity monoclonal antibodies. This proprietary polymerase mix is highly resistant to PCR inhibitors and provides an extremely stringent automatic hot-start allowing reaction assembly, and temporary storage, at room temperature prior to PCR amplification. PerfeCTa qPCR ToughMix delivers exceptional performance with either fast or conventional PCR cycling protocols.
UNG containing versions are blended with Uracil N-glycosylase to eliminate potential post-PCR carryover contamination associated with routine molecular testing.
Details
Single-tube, 2X concentrated reagent containing:
- Reaction buffer with optimized concentrations of molecular-grade MgCl2, dATP, dCTP, dGTP, dTTP.
- AccuStart II Taq DNA Polymerase.
- Inert AccuVue dye.
- Proprietary enzyme stabilizers and performance-enhancing additives.
- Titrated reference dye (if applicable).
Instrument Compatibility
ROX
- Applied Biosystems 5700
- Applied Biosystems 7000
- Applied Biosystems 7300
- Applied Biosystems 7700
- Applied Biosystems 7900
- Applied Biosystems 7900HT
- Applied Biosystems 7900 HT Fast
- Applied Biosystems StepOne™
- Applied Biosystems StepOnePlus™
Low ROX
- Applied Biosystems 7500
- Applied Biosystems 7500 Fast
- Stratagene Mx3000P®
- Stratagene Mx3005P™
- Stratagene Mx4000™
- Applied Biosystems ViiA 7
- Applied Biosystems QuantStudio™ (all models)
- Douglas Scientific IntelliQube®
No ROX
- Quantabio Q
- BioRad CFX
- Roche LightCycler 480
- QIAGEN Rotor-Gene Q
- Agilent AriaMx
- Azure Cielo™
- Analytik Jena qTower
- Analytik Jena qTOWERiris
- Other
Bio-Rad iCycler iQ systems
- BioRad iCycler iQ™
- BioRad MyiQ™
- BioRad iQ™5
Performance Data
Comparison of PerfecTa qPCR ToughMix Sensitivity in the presence of high presence of inhibitors against other enzyme manufacturers
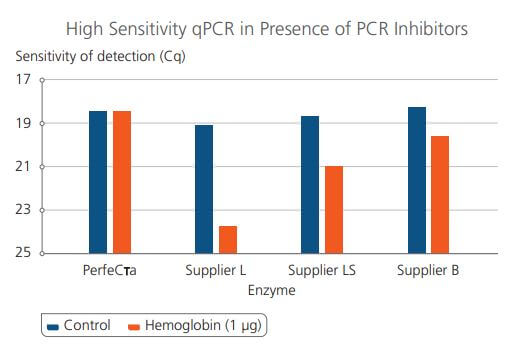
Comparison of PerfecTa qPCR ToughMix Sensitivity in the presence of high presence of inhibitors against other enzyme manufacturers
Comparison of PerfecTa qPCR ToughMix Sensitivity in the presence of high presence of inhibitors against other enzyme manufacturers
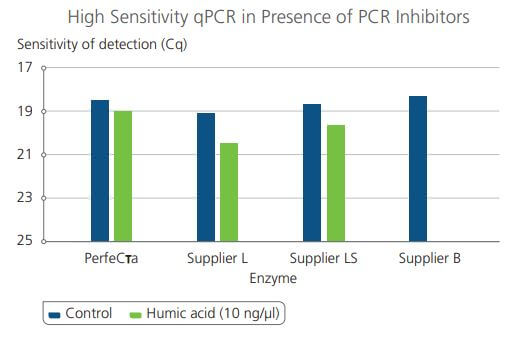
Comparison of PerfecTa qPCR ToughMix Sensitivity in the presence of high presence of inhibitors against other enzyme manufacturers
Resources
Customer Profile Stories
Flyers
Product Manuals
Application Notes
CofA (PSFs)
SDSs
Publications
Customer Product Reviews
| 5 star | 100% | |
| 4 star | 0% | |
| 3 star | 0% | |
| 2 star | 0% | |
| 1 star | 0% |
 PerfeCTa qPCR ToughMix
PerfeCTa qPCR ToughMix

Easy to use, about as sensitive as NEB’s LUNA RT-qPCR reagents
Although a small sample size was tested, the ToughMix Master Mix resulted in better sensitivity on our Campylobacter assay from fecal specimens (only 4 samples were tested), there appeared to be approximately 1 CT value decrease compared to our normal master mix TaqMan Fast Virus 1-Step Master Mix. Additionally, the PCR run time was able to be reduced by approximately 30 minutes and this product is cheaper. I plan on discussing with my PI about switching the SOP. Thank you!
I compared PerfeCTa qPCR toughMix L-ROX 250R with Luna® Universal Probe qPCR Master Mix (NEB) on the amplification of dsrA (sulfite reductase). PerfeCTa qPCR ToughMix L-ROX 250R is much better than Luna® Universal Probe qPCR Master Mix.
This is a good product that did the job it should do. The sensitivity is almost the same as the one I am currently using. I do not mind switching to it if it can be optimized to match or beat the others for the sensitivity.