
sparQ PureMag Beads
- High recovery of DNA fragments greater than 100 bp
- Efficient removal of unwanted components from adapter ligation and PCR reactions
- Consistent single or double-sided size selection
- Seamless integration into existing NGS workflows with little or no protocol change
- Easy-to-use and compatible with manual processing or automated liquid handling robots
- Cost effective alternative to AMPure® XP with equivalent performance
sparQ PureMag Beads
Description
sparQ PureMag Beads is a fast and reliable nucleic acid purification system for reaction cleanup and size selection in Next Generation Sequencing (NGS) workflows. Based on the reversible nucleic acid-binding properties of magnetic beads, this product can be used to quickly remove primers, primer-dimers, unincorporated nucleotides, salts, adapters and adapter-dimers from NGS library prep reactions to improve downstream sequencing performance. sparQ PureMag Beads allows excellent recovery of fragments greater than 100 bp without centrifugation or filtration. Consistent and reliable size selection can be achieved by simply adjusting the beads to sample ratio. This product is designed for both manual and automated processing, allowing seamless integration into existing workflows.
Details
Performance Data
Efficient recovery of DNA equivalent to AMPure XP
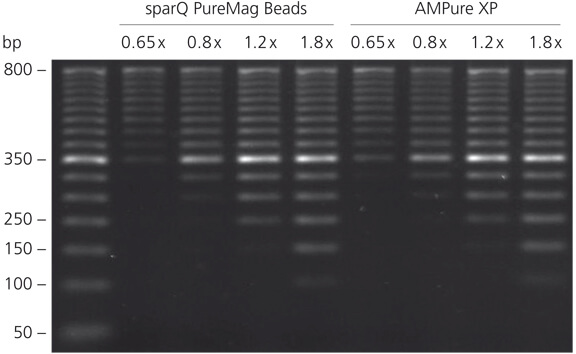
sparQ PureMag Beads show equivalent performance to AMPure XP for DNA purification. 50 bp DNA ladder was purified with sparQ PureMag Beads and AMPure XP at different beads to DNA ratios and analyzed on 2% agarose gel.
Highly reproducible purification across a range of inputs
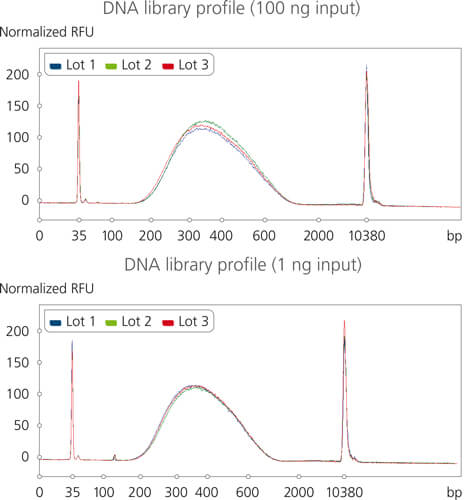
Highly reproducible DNA library profiles were achieved using different lots of sparQ PureMag Beads and a broad range of input amount. Libraries were prepared with sparQ DNA Library Prep Kit from 100 ng and 1 ng of fragmented microbial genomic DNA. sparQ PureMag Beads were used post adapter ligation and PCR amplification to effectively remove adapter-dimers and primer-dimers.
Bioanalyzer trace of fragmented human genomic DNA prior and post double-sided size selection
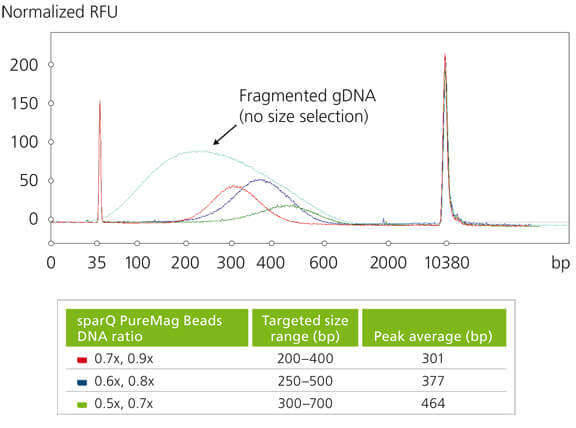
Electropherogram of fragmented human genomic DNA prior and post double-sided size selection. Different sparQ PureMag Beads to DNA ratios were used to achieve various targeted size range.
sparQ PureMag beads workflow
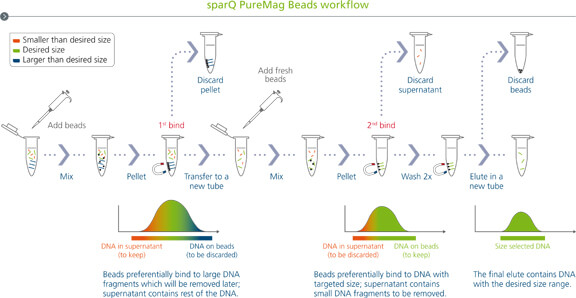
Double-sided size selection is used to remove smaller and larger fragments from either side of the desired region. The fragment size can be easily adjusted to suit the application by manipulating the sparQ PureMag Beads to DNA volumetric ratio.
Resources
Customer Profile Stories
Flyers
Product Manuals
Application Notes
Brochures
CofA (PSFs)
SDSs
Publications
Customer Product Reviews
| 5 star | 72% | |
| 4 star | 9% | |
| 3 star | 18% | |
| 2 star | 0% | |
| 1 star | 0% |
 sparQ PureMag Beads
sparQ PureMag Beads


Worked as expected for magnetic cleanup beads.
We saw no difference in the performance of the SparQ beads vs. the Ampure beads despite them being much cheaper. We tested their ability to bind tRNAs and they were not able to (which is what we wanted and expected). This is the same as what we observed for Ampure beads. We haven’t tested them yet on their ability to bind ~330 nt chimeric RNA/DNA molecule which we make in library preparations, but we are confident they will work given what we’ve seen so far.
Easy and straight-forward to use. Very approachable method for students leaning to purify DNA.
Recovery was better than AMPure XP beads. Also, these were sitting at room temperature (accidentally) for about a month before I used them.
Great product!