
qScript Flex cDNA Synthesis Kit
- Separate components for user-defined RT priming method, (oligo-dT, random hexamer, or GSP)
- Highly sensitive first-strand cDNA synthesis of RNA sequences ≤ 12kb for quantitative and conventional two-step RT-PCR
- Broad linear dynamic detection range with limiting (10pg) or plentiful (4ug) total RNA samples
- Maximize cDNA yield with proprietary Priming Enhancer additive
qScript Flex cDNA Kit is intended for molecular biology applications. This product is not intended for the diagnosis, prevention or treatment of a disease.
qScript Flex cDNA Synthesis Kit
Description
The qScript Flex cDNA Synthesis Kit is configured in separate components for supporting multiple RT priming strategies. The kit provides optimized, flexible components for priming with oligo-dT, random hexamers, gene-specific primer (GSP), or any combination thereof. This unique 5X master mix of buffer, magnesium, stabilizers and dNTPs simplifies reaction assembly and ensures robust and reproducible synthesis of first strand product from 10 pg to 4µg of total RNA or purified polyA+ RNA template. The resulting cDNA product is directly compatible with current real-time RT-PCR methods or end-point RT-PCR. Length of cDNA product is dependent upon priming strategy and quality of the RNA template. Oligo dT or GSP can be used for two-step RT-PCR of RNA targets up to 12 kb. Random primer is suitable for RT-PCR of RNA targets less than 1 kb. Any of the three priming methods are suitable for real-time quantitative RT PCR (RT-qPCR).
For one-step RT-PCR of targets less than 5 kb, see our qScript XLT 1-Step RT-PCR Kit
Details
- Master mix, 5X concentrated solution of optimized reaction buffer, magnesium, dNTP blend, proprietary stabilizers and additives
- Oligo dT(20), 10X concentrated with Priming Enhancer
- Random Primer, 10X concentrated with Priming Enhancer
- Priming Enhancer, 10X concentrated
- Stabilized qScript Reverse Transcriptase with ribonuclease inhibitor protein (RIP)
- Nuclease-free water
Performance Data
Linearity
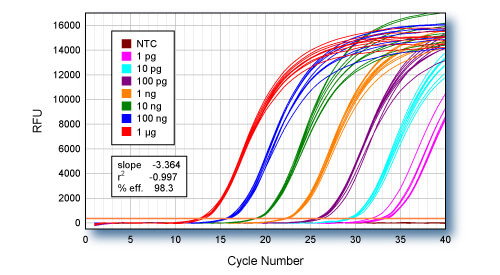
qScript Flex cDNA Synthesis Kit with oligo dT primer was used for first-strand cDNA synthesis using log-fold serial dilutions of HeLa cell total RNA from 1 µg to 1 pg. Eight replicate cDNA reactions were performed for each input quantity of RNA. 1/10th of each first-strand reaction was used for qPCR of the ACTB gene using PerfeCTa™ SYBR Green SuperMix.
Resources
Product Manuals
Application Notes
CofA (PSFs)
SDSs
Publications
Customer Product Reviews
| 5 star | 100% | |
| 4 star | 0% | |
| 3 star | 0% | |
| 2 star | 0% | |
| 1 star | 0% |
 qScript Flex cDNA Synthesis Kit
qScript Flex cDNA Synthesis Kit

I appreciate both the price and inclusion of RNAse inhibitor in the qScript Flex, and in my miRNA assay development I appreciate the ability to have no RT controls with the separate addition of the enzyme.