AccuStart II PCR Genotyping Kit
- Simplified, completely reagent-based system requires minimal pipetting skill
- Premixed electrophretic mobility loading dye reduces chances for post-PCR cross contamination
- Stabilized 2X PCR SuperMix enables convenient room-temperature setup and is unaffected by repetitive freeze-thaw (>20X)
- High-yielding, ultrapure modified Taq DNA polymerase delivers robust, reliable duplex assay performance
- Stringent, ultrapure antibody hotstart ensures sensitive and specific target amplification
AccuStart II PCR Genotyping Kit is intended for molecular biology applications. This product is not intended for the diagnosis, prevention or treatment of a disease.
AccuStart II PCR Genotyping Kit
Description
The AccuStart II Genotyping Kit is a complete reagent kit designed to support conventional, end-point PCR-based screening of transgenic animal models commonly used in life science research and is validated for use with mouse, fish, or insect tissue specimens. It combines a rapid, 2-component DNA extraction reagent with a user-friendly 2X concentrated PCR SuperMix with loading dye for seamless gel electrophoresis analysis. qPCR-grade genomic DNA template is obtained with minimal extraction volumes (100uL) and can be carried out in 30-minutes on a standard PCR thermal cycler.
Details
- Extracta® DNA Prep for PCR (95091-02)
- Extraction Reagent
- Stabilization Buffer
- AccuStart II GelTrack PCR SuperMix (95136-500)
- 2X concentrated SuperMix containing optimized concentrations of molecular-grade MgCl2, dNTP blend, AccuStart II Taq DNA Polymerase, reaction buffer, stabilizers and electrophoretic mobility dyes (4kb & 50bp).
Performance Data
AccuStart II Results more specific
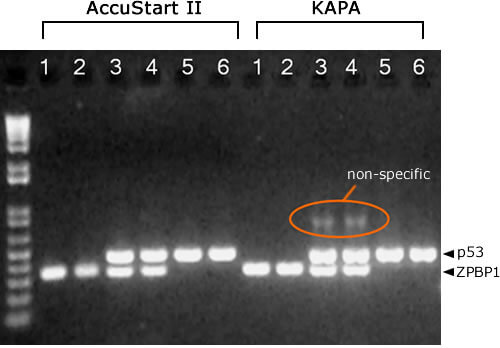
KAPA PCR mix requires more optimization to reduce non-specific amplification products in multiplex PCR reactions. Lanes: 1 = mouse 1, ZPBP1. 2 = mouse 2, ZPBP1. 3 = mouse 1, ZPBP1/p53. 4 = mouse 2, ZPBP1/p53. 5 = mouse 1, p53. 6 = mouse 2, p53
Resources
Product Manuals
CofA (PSFs)
SDSs
Publications
Customer Product Reviews
| 5 star | 75% | |
| 4 star | 25% | |
| 3 star | 0% | |
| 2 star | 0% | |
| 1 star | 0% |

The AcuuStart II Mouse Genotyping Kit 100R has worked excellent in our current mouse genotyping protocols. The overall sensitivity was improved immensely. Additionally, the kit decreased the amount of time needed to prepare the DNA for PCR amplification.
We have previously tried the extraction system, and we are very satisfied with the system. This time, we tested the efficacy of the PCR system. The PCR results were robust, and the procedure was straightforward.
The speed and ease of this for extracting DNA from fish fin clips is amazing! We’re getting clean qPCR (genotyping) results and haven’t had a sample fail yet! Best yet is that it only takes 30min, compared to our previous workflow which took a few hours and had ~80% success rate.