Extracta DBS
- Optimized for DNA extraction from dried blood spot punches
- Single reagent for PCR-ready DNA in 30 minutes
- Maximized assay sensitivity, lower Cq values, when combined with Quantabio ToughMix®
- Compatible with high-throughput automation for PCR, qPCR and NGS applications
Extracta DBS is intended for molecular biology applications. This product is not intended for the diagnosis, prevention or treatment of a disease.
Extracta DBS
Description
Extracta DBS is a ready-to-use DNA extraction reagent for rapid and efficient recovery of PCR-ready DNA from dried blood spots (DBS) on Guthrie cards or Whatman 903 filter paper. This patented single-solution process produces DNA eluates that are substantially free of PCR inhibitors and compatible with a variety of end-point PCR, real-time PCR and Next Generation Sequencing (NGS) or Sanger Sequencing (1-3) reagents. Application of Extracta DBS with PerfeCTa® qPCR ToughMix or PerfeCTa MultiPlex qPCR ToughMix enables accurate and reproducible quantification of DNA sequences in blood using TaqMan® hydrolysis probe real-time qPCR.
Combine with ToughMix and maximize qPCR assay sensitivity
Extracta DBS is the perfect match with Quantabio ToughMix for sensitive and precise target quantification. The crude extraction combined with ToughMix, a Quantabio master mix that is tolerant to common PCR inhibitors, results in higher DNA yields independent of DNA sample inputs and qualities to enable accurate detection and high sensitivity even with low copy targets.
Reliable, Consistent Lot-to-Lot Performance
Low amounts and quality of DNA recovered from dried blood spots commonly restrict the utilization of DNA. To overcome these limitations, Extracta DBS increases the yield and quality allowing for efficient and reliable recovery of DNA from dried blood spots. Manufactured under stringent quality standards, Extracta DBS ensures uniform lot-to-lot performance resulting in reliable reproducibility in combination with Quantabio ToughMix.
Details
Extracta DBS is stable for up to 2 years at 2-8°C. Please consult the product box label for expiration date information.
Performance Data
TREC Control Sample
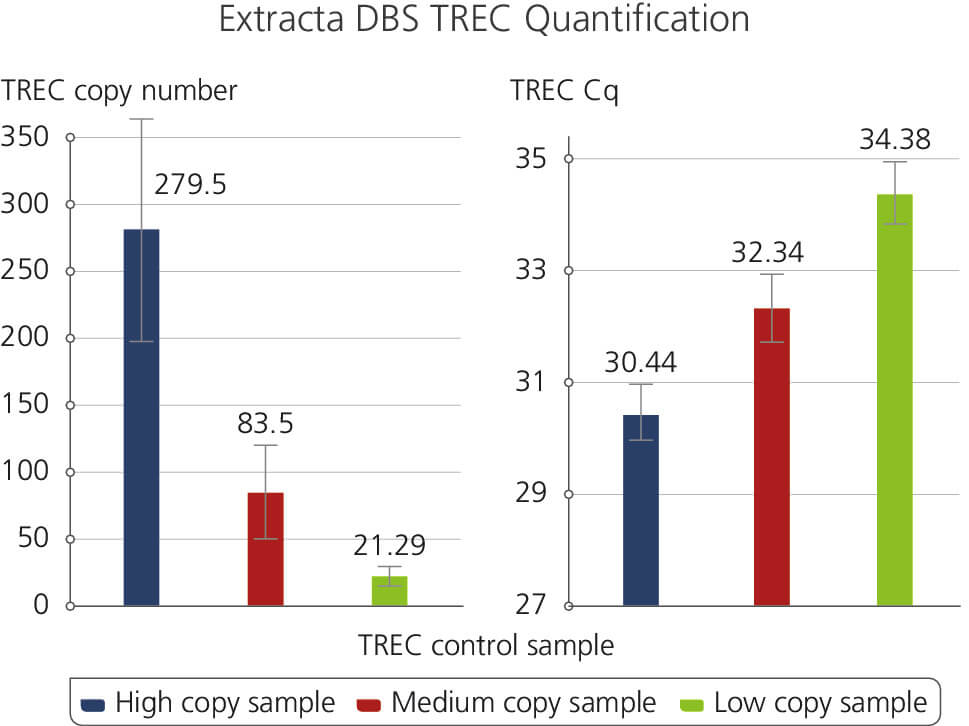
Illustrates the results of a T-cell Receptor Excision Circles (TREC) Assay using Extracta DBS and PerfeCTa ToughMix. Samples were generated using dried blood spot punches following the Extracta DBS protocol and used subsequently for quantification of T-cell Receptor Excision Circles. The samples are representatives of High, Medium and Low TREC copy numbers along with the corresponding Cq values.
Different Extracta DBS preparation
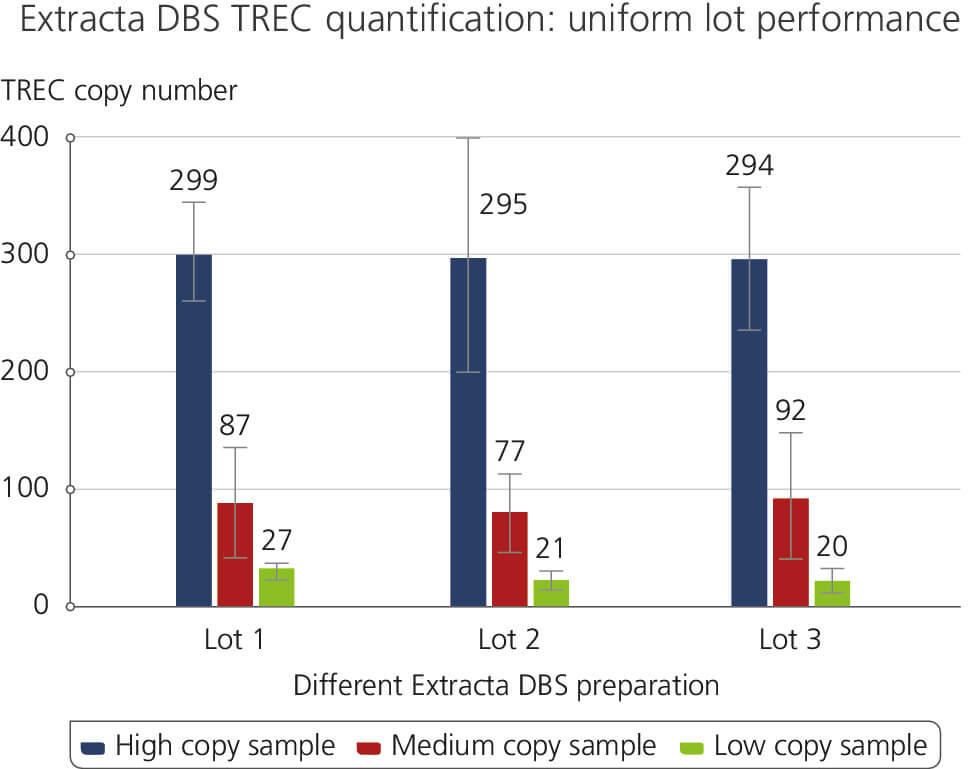
This figure demonstrates consistent lot-to-lot performance in a TREC quantification assay using genomic DNA extracted from dried blood spots. Lot-to-lot performance was tested for High, Medium and Low copy number TREC samples. The results highlight the reliability and reproducibility across various product lots which are attributed to Quantabio’s high manufacturing and production standards under ISO 13485.
Fast, single solution extraction workflow in 30 min
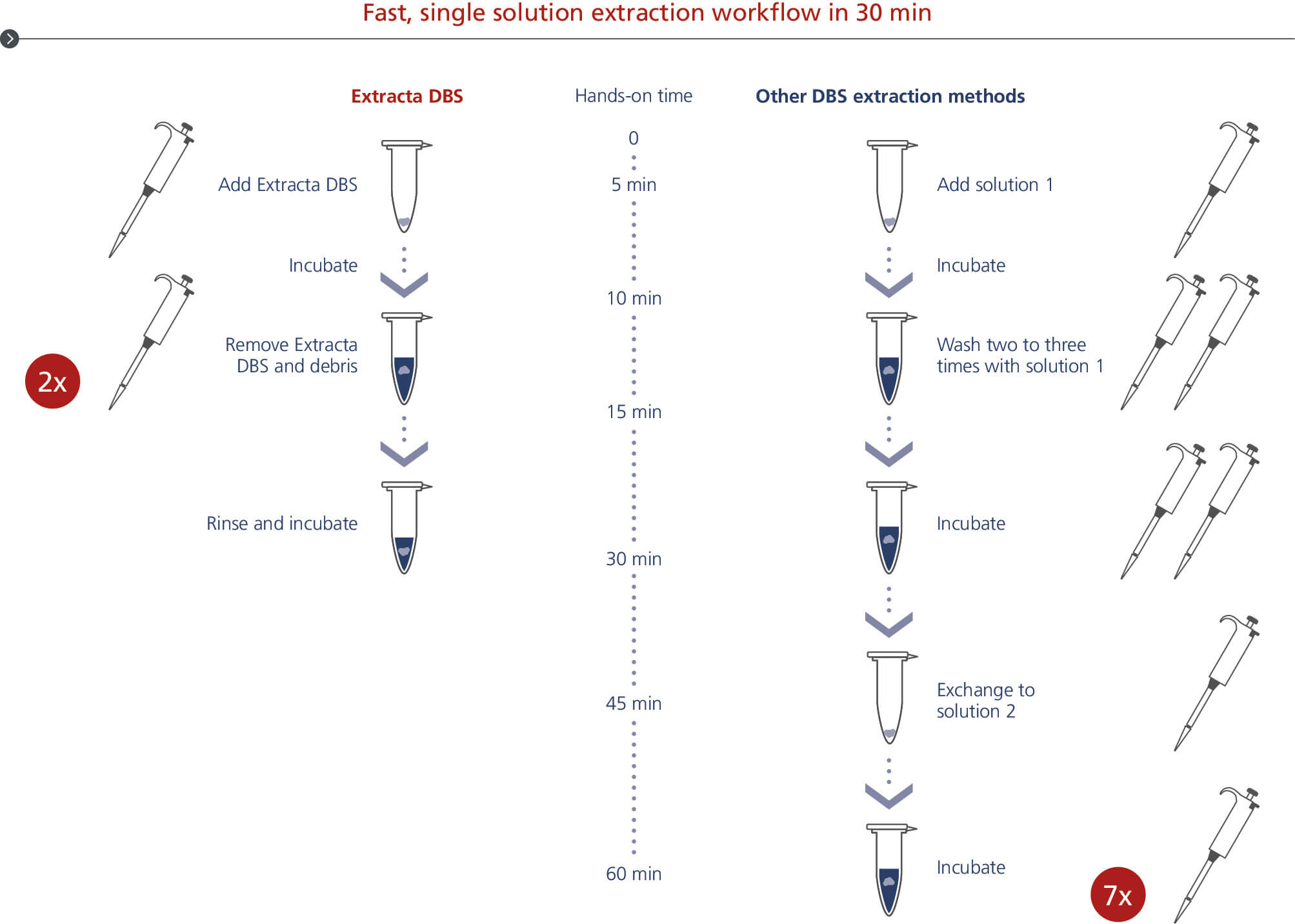
Extracta DBS DNA extraction from dried blood spots workflow. PCR-ready DNA is eluated from dried blood spots with less handling in 30 minutes.
Documents & Downloads
Customer Product Reviews
| 5 star | 50% | |
| 4 star | 50% | |
| 3 star | 0% | |
| 2 star | 0% | |
| 1 star | 0% |
 Extracta DBS
Extracta DBS

I did not have success with your extraction buffer but I believe it has to do with the FTA cards I was extracting DNA. It is not recommended not to elude DNA from FTA Classic cards and these cards are different than Gutherie cards as they have proprietary lysing agents and preservation chemicals stabilize DNA and recommend washing with their proprietary rinse buffer. I tried Extracta DBS with and without washing with the FTA rinse buffer but I ran out of your mastermix to try and optimize the pcr.
We compared Extracta DBS with current method (proteinase K and spin column) using PCR as a downstream tool . The Extracta DBS was superior to this method for genomic DNA. However, the current method was superior for recovering low copies of viral DNA.