AccuStart Genotyping ToughMix
- Optimized buffer chemistry destabilizes single base-pair mismatch probes, providing superior allelic discrimination and improved cluster separation for critical, single-nucleotide polymorphism (SNP) detection assays
- Sensitive, precise detection with ultrapure AccuStart II enzyme technology – maximum-yielding Taq DNA polymerase mutant controlled by stringent, multi-epitope antibody hotstart
- ToughMix reagent technology neutralizes a broad spectrum of PCR inhibitors common in plant and animal tissues, environmental sources, clinical specimens and complex food matrices
- Easy-to-use 2x concentrated SuperMix with AccuVue plate loading dye and pre-blended passive reference dye simplifies reaction setup
- Supports efficient vortex mixing with proprietary anti-foaming technology
Genotyping ToughMix is intended for molecular biology applications. This product is not intended for the diagnosis, prevention or treatment of a disease.
Genotyping ToughMix ROX
Genotyping ToughMix Low ROX
Description
Genotyping ToughMix is a 1-tube qPCR SuperMix reagent compatible with all dual-label (hydrolysis) probe chemistries for both fast and conventional PCR cycling protocols or instruments. This proprietary formulation has been rigorously optimized to destabilize single base-pair mismatches to ensure precise allelic discrimination and cluster separation with SNP detection assays. The reagent is provided as a 2x concentrated ready-to-use reaction cocktail that contains all required reaction components, except primers, probe(s), and DNA template. Inert AccuVue plate loading dye helps to minimize pipette error and provides visual confirmation of thorough mixing.
Details
Single-tube, 2X concentrated reagent containing:
- Reaction buffer with optimized concentrations of molecular-grade MgCl2, dATP, dCTP, dGTP, dTTP.
- AccuStart II Taq DNA Polymerase.
- Inert AccuVue dye.
- Proprietary enzyme stabilizers and performance-enhancing additives.
- Titrated reference dye (if applicable).
Instrument Compatibility
ROX
- Applied Biosystems 5700
- Applied Biosystems 7000
- Applied Biosystems 7300
- Applied Biosystems 7700
- Applied Biosystems 7900
- Applied Biosystems 7900HT
- Applied Biosystems 7900 HT Fast
- Applied Biosystems StepOne™
- Applied Biosystems StepOnePlus™
Low ROX
- Applied Biosystems 7500
- Applied Biosystems 7500 Fast
- Stratagene Mx3000P®
- Stratagene Mx3005P™
- Stratagene Mx4000™
- Applied Biosystems ViiA 7
- Applied Biosystems QuantStudio™ (all models)
- Douglas Scientific IntelliQube®
No ROX
- Quantabio Q
- BioRad CFX
- Roche LightCycler 480
- QIAGEN Rotor-Gene Q
- Agilent AriaMx
- Azure Cielo™
- Analytik Jena qTower
- Analytik Jena qTOWERiris
- Other
Bio-Rad iCycler iQ systems
- BioRad iCycler iQ™
- BioRad MyiQ™
- BioRad iQ™5
Performance Data
Comparison to conventional master mixes
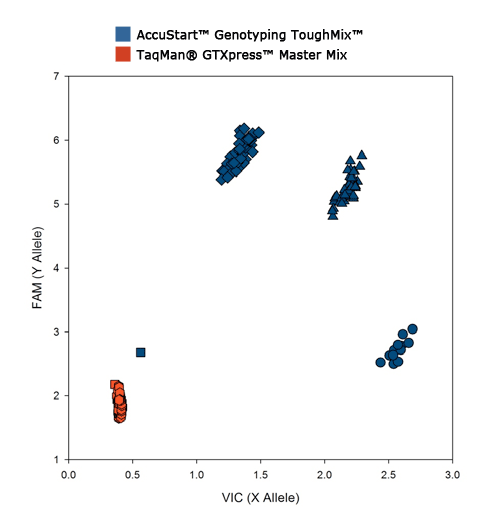
In the presence of a common PCR inhibitor, humic acid (50ng/l), the competitors system is completely shut down while ToughMix delivers robust, accurate results
Comparison to conventional master mixes
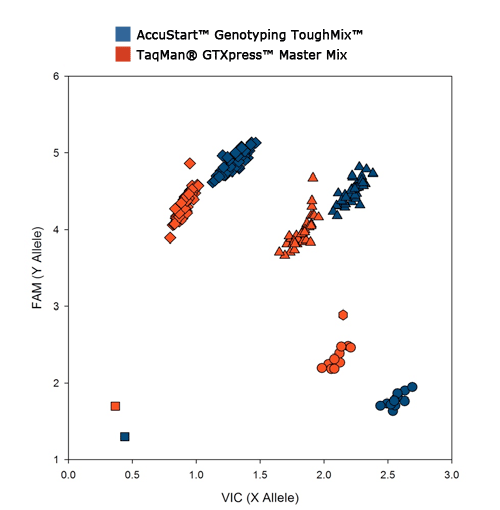
AccuStart Genotyping ToughMix stands up to the challenge where other genotyping master mixes fall apart. ToughMix can be used with clean templates where it generates higher fluorescent signal and tighter clusters than the leading competitors mix.
Resources
Flyers
Product Manuals
Application Notes
CofA (PSFs)
SDSs
Publications
Customer Product Reviews
| 5 star | 0% | |
| 4 star | 100% | |
| 3 star | 0% | |
| 2 star | 0% | |
| 1 star | 0% |
 AccuStart Genotyping ToughMix
AccuStart Genotyping ToughMix

more dye to improve the visualization of the mastermix.