PerfeCTa SYBR® Green FastMix
- 2x concentrated reagents minimize pipetting steps, simplify reaction assembly and improve accuracy
- Superior assay sensitivity and specificity with ultrapure AccuStart enzyme technology – maximum-yielding Taq DNA polymerase mutant controlled by stringent, multi-epitope antibody hot start
- Supports efficient vortex mixing with proprietary anti-foaming formulation
- FastMix formulation supports both fast and standard thermal cycling conditions
PerfeCTa SYBR Green FastMix is intended for molecular biology applications. This product is not intended for the diagnosis, prevention or treatment of a disease.
PerfeCTa SYBR Green FastMix for IQ
PerfeCTa SYBR Green FastMix
PerfeCTa SYBR Green FastMix ROX
PerfeCTa SYBR Green FastMix Low ROX
Description
PerfeCTa SYBR® Green FastMix is a 2X concentrated, ready-to-use reaction cocktail that contains all components, except primers and DNA template. This rigorously optimized master mix contains of proprietary buffer technology, stabilizers and AccuFast Taq DNA polymerase to deliver maximum assay precision, sensitivity, and PCR efficiency for accelerated or conventional thermal cycling conditions for SYBR Green detection. Dye-based detection methods are critically dependent on highly specific amplification because dsDNA dyes will bind to any amplicon, including off-target primer elongation and primer dimerization. AccuFast hot start Taq DNA polymerase contains a proprietary mixture of ultra-pure monoclonal antibodies that stringently suppress primer elongation prior to the initial PCR denaturation step and allows for setup and multi-day storage at ambient room temperature prior to thermal cycling. AccuFast provides rapid release of fully active enzyme to support accelerated thermal cycling conditions.
Details
Single-tube, 2X concentrated reagent containing:
- Reaction buffer with optimized concentrations of molecular-grade MgCl2, dATP, dCTP, dGTP, and dTTP.
- AccuStart II Taq DNA Polymerase
- SYBR Green I dye
- Proprietary enzyme stabilizers and performance-enhancing additives.
- Titrated reference dye (if applicable)
Instrument Compatibility
ROX
- Applied Biosystems 5700
- Applied Biosystems 7000
- Applied Biosystems 7300
- Applied Biosystems 7700
- Applied Biosystems 7900
- Applied Biosystems 7900HT
- Applied Biosystems 7900 HT Fast
- Applied Biosystems StepOne™
- Applied Biosystems StepOnePlus™
Low ROX
- Applied Biosystems 7500
- Applied Biosystems 7500 Fast
- Stratagene Mx3000P®
- Stratagene Mx3005P™
- Stratagene Mx4000™
- Applied Biosystems ViiA 7
- Applied Biosystems QuantStudio™ (all models)
- Douglas Scientific IntelliQube®
No ROX
- Quantabio Q
- BioRad CFX
- Roche LightCycler 480
- QIAGEN Rotor-Gene Q
- Agilent AriaMx
- Azure Cielo™
- Analytik Jena qTower
- Analytik Jena qTOWERiris
- Other
Bio-Rad iCycler iQ systems
- BioRad iCycler iQ™
- BioRad MyiQ™
- BioRad iQ™5
Performance Data
PerfeCTa SYBR Green FastMix comparison to SYBR GreenER qPCR SuperMix
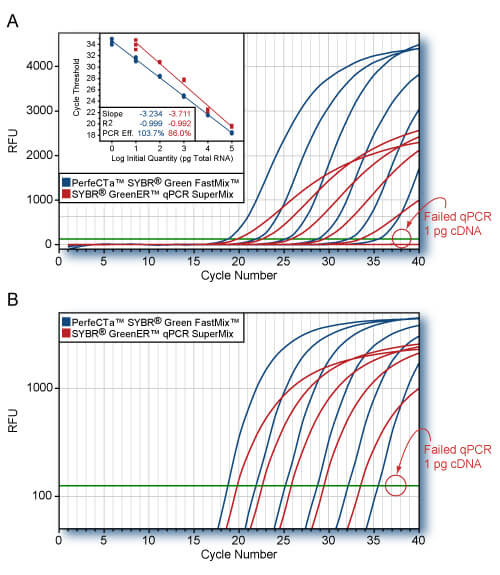
RNA-specific adenosine deaminase (ADAR) was amplified from log-fold dilutions of total HeLa cell cDNA (100 ng to 1 pg) using PerfeCTaTM; SYBR Green FastMix or the SYBR GreenER qPCR SuperMix (Invitrogen) according to each manufacturers protocol. Averaged plots for quadruplicate reactions for each input quantity are shown. Replicate CT values are shown on the standard curve (Panel A, inset). Cycling conditions: Invitrogenn: 95°C, 10 min followed by 40 cycles of 95°C, 10s; 60°C, 60s; PerfeCTaTM; SYBR Green FastMix: 95°C, 20s followed by 40 cycles of 95°C, 1s; 60°C, 20s. PerfeCTaTM SYBR Green FastMix amplified the ADAR gene with higher efficiency and greater sensitivity. All replicate reactions for SYBR GreenER qPCR SuperMix failed to amplify ADAR from 1 pg of cDNA.
PerfeCTa SYBR Green FastMix Comparison to SYBR PreMix Ex Taq
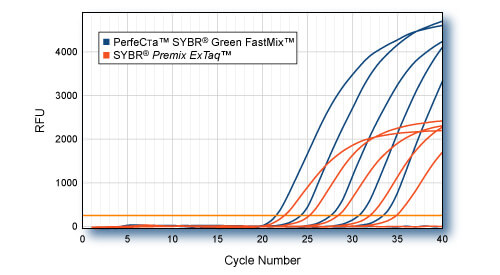
RNA-specific adenosine deaminase (ADAR) was amplified from log-fold dilutions of total HeLa cell cDNA (100 ng to 10 pg) using PerfeCTaTM SYBR Green FastMix or SYBR PreMix Ex TaqTM (Takara) according to each manufacturers protocol. Averaged plots for quadruplicate reactions for each input quantity are shown. PerfeCTaTM; SYBR Green FastMix produces higher fluorescent signal and detection of equal target amounts at earlier Cts. Cycling conditions for both kits: 95°C, 20s followed by 40 cycles of 95°C, 1s; 60°C, 20s
PerfeCTa SYBR Green FastMix Comparison to DyNAmo Flash SYBR Green PCR Kit
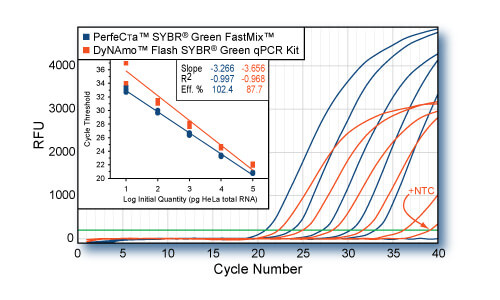
RNA-specific adenosine deaminase (ADAR) was amplified from log-fold dilutions of total HeLa cell cDNA (100 ng to 10 pg) using PerfeCTa SYBR Green FastMix or the DyNAmo Flash SYBR Green PCR Kit (Finnzymes) according to each manufacturers protocol. Averaged plots for quadruplicate reactions for each input quantity are shown. Fusion of DNA-binding peptide to Tbr DNA pol results in lower specificity of the DyNAmo kit which is evident in false positive results for no template control (NTC) reactions. Chemically modified polymerase produces delayed Cts and lower signal strength compared to AccuStart™ Taq. Cycling conditions: Finnzymes: 95°C, 7 min followed by 40 cycles of 95°C, 10s; 60°C, 20s PerfeCTaTM SYBR Green FastMix: 95°C, 20s followed by 40 cycles of 95°C, 1s; 60°C, 20s
Resources
Customer Profile Stories
Flyers
Product Manuals
Application Notes
CofA (PSFs)
SDSs
Publications
Customer Product Reviews
| 5 star | 80% | |
| 4 star | 17% | |
| 3 star | 0% | |
| 2 star | 0% | |
| 1 star | 2% |
 PerfeCTa SYBR® Green FastMix
PerfeCTa SYBR® Green FastMix

This mix was super simple to use. I didn’t even have to change my usual protocol to accomodate for the change.
Sara, an undergraduate student, used the PerfeCTa SYBR Green FastMix on the QuantaBio qPCR instrument. We were easily able to figure out and make master mixes. The fast cycling results were impressive! Thank you!
We have previously been using the BioRad supermix for our CFX384 machine qPCR reagents. We found the PerfeCTa mix has much higher sensitivity than the BioRad supermix. We are impressed with the price and quality and will be switching products!
We found that the PerfeCTa SYBR FastMix performed significantly better than our current supplier.